Luay Nakhleh
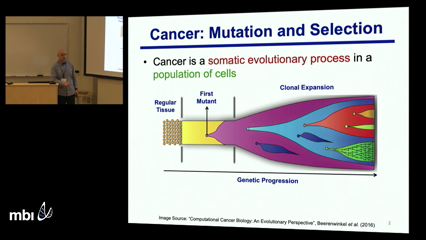
Intra-tumor heterogeneity, as caused by a combination of mutation and selection, poses significant challenges to the diagnosis and treatment of cancer. Resolving this heterogeneity to identify the tumor cell populations (clones) and delineate their evolutionary history is of critical importance in improving cancer diagnosis and therapy. This heterogeneity can be readily elucidated and understood through the reconstruction of the clonal genotypes and evolutionary history of the tumor cells. Recently introduced single-cell DNA sequencing (SCS) technologies promise to provide the appropriate type of data for resolving intra-tumor heterogeneity. However, inherent technical errors in SCS datasets, due to allelic dropout, cell doublets and coverage non-uniformity, significantly complicate these tasks.
In this talk, I will first describe a maximum likelihood method for inferring tumor trees from SCS genotype data with potentially erroneous and missing entries, under a finite-sites model of evolution. I will then describe a non-parametric Bayesian method that simultaneously reconstructs the clonal populations as clusters of single cells, mutations associated with each clone, and the genealogical relationships between the clonal populations. I will demonstrate the performance of the methods on both synthetic and real data sets.
This is collaborative work with Hamim Zafar, Anthony Tzen, Ken Chen, and Nicholas Navin.
 Luay NakhlehIntra-tumor heterogeneity, as caused by a combination of mutation and selection, poses significant challenges to the diagnosis and treatment of cancer. Resolving this heterogeneity to identify the tumor cell populations (clones) and delineate their evolutionary history is of critical importance in improving cancer diagnosis and therapy. This heterogeneity can be readily elucidated and understood through the reconstruction of the clonal genotypes and evolutionary history of the tumor cells. Recently introduced single-cell DNA sequencing (SCS) technologies promise to provide the appropriate type of data for resolving intra-tumor heterogeneity. However, inherent technical errors in SCS datasets, due to allelic dropout, cell doublets and coverage non-uniformity, significantly complicate these tasks.
Luay NakhlehIntra-tumor heterogeneity, as caused by a combination of mutation and selection, poses significant challenges to the diagnosis and treatment of cancer. Resolving this heterogeneity to identify the tumor cell populations (clones) and delineate their evolutionary history is of critical importance in improving cancer diagnosis and therapy. This heterogeneity can be readily elucidated and understood through the reconstruction of the clonal genotypes and evolutionary history of the tumor cells. Recently introduced single-cell DNA sequencing (SCS) technologies promise to provide the appropriate type of data for resolving intra-tumor heterogeneity. However, inherent technical errors in SCS datasets, due to allelic dropout, cell doublets and coverage non-uniformity, significantly complicate these tasks.